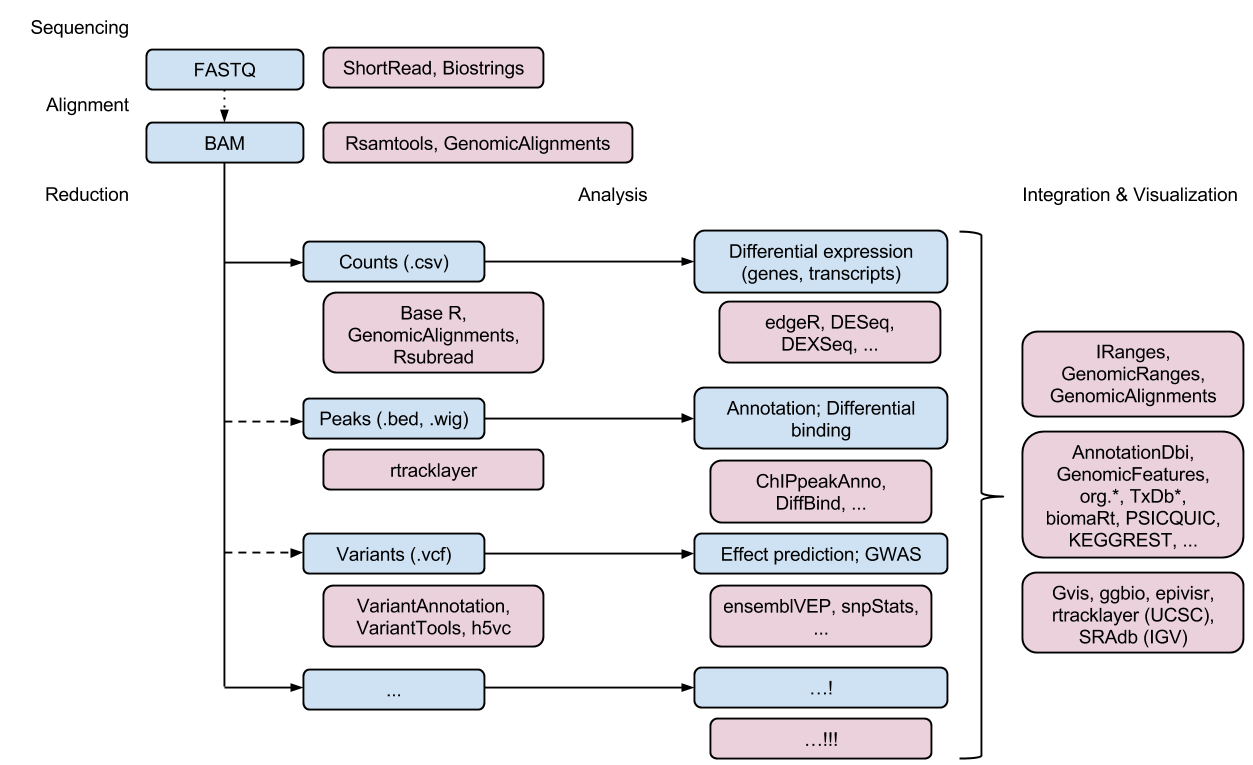
High-throughput sequence workflow
Bioconductor
Analysis and comprehension of high-throughput genomic data
Statistical analysis designed for large genomic data
Interpretation: biological context, visualization, reproducibility
Support for all high-throughput technologies
- Sequencing: RNASeq, ChIPSeq, variants, copy number, ...
- Microarrays: expression, SNP, ...
- Flow cytometry, proteomics, images, ...
Bioconductor cheat sheet https://github.com/mikelove/bioc-refcard
Bioconductor by the numbers
Project started in 2002
Built on and in R, the open source software platform for data science
An estimated 2,000,000 users worldwide
More than 50,000 unique downloads per month
More than 22,000 PubmedCentral citations
Bioconductor Release: 1,974 biomedical and omics data science software packages (02-20-2021)
Receiving submissions of 3-6 new packages per week
Bioconductor: Software for orchestrating high-throughput biological data analysis by Sean Davis
Reference manuals, vignettes
All user-visible functions have help pages, most with runnable examples
'Vignettes' an important feature in Bioconductor -- narrative documents illustrating how to use the package, with integrated code
Example:
AnnotationHublanding page, AnnotationHub HOW TO's vignette illustrating some fun use cases
https://bioconductor.org/packages/AnnotationHub/
Bioconductor classes
Bioconductor makes extensive use of classes to represent complicated data types
- The core components: classes, generic functions and methods
- The S4 class system is a set of facilities for object-oriented programming
Classes foster interoperability - many different packages can work on the same data - but can be a bit intimidating
Formal S4 object system
Often a class is described on a particular home page, e.g.,
?GRanges, and in vignettes, e.g.,vignette(package="GenomicRanges"),vignette("GenomicRangesIntroduction")Many methods and classes can be discovered interactively , e.g.,
methods(class="GRanges")to find out what one can do with aGRangesinstance, andmethods(findOverlaps)for classes that thefindOverlaps()function operates onIn more advanced cases, one can look at the actual definition of a class or method using
getClass(),getMethod()Getting help:
?findOverlaps,<tab>to select help on a specific method,?GRanges-classfor help on a class.
High-throughput sequence data
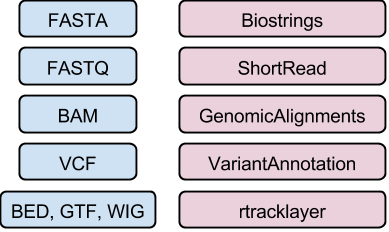
DNA/amino acid sequences: FASTA files
- The
Biostringspackage is used to represent DNA sequences, with many convenient sequence-related functions, e.g.,?consensusMatrix.
Input & manipulation, FASTA file example:
>NM_078863_up_2000_chr2L_16764737_f chr2L:16764737-16766736gttggtggcccaccagtgccaaaatacacaagaagaagaaacagcatcttgacactaaaatgcaaaaattgctttgcgtcaatgactcaaaacgaaaatg...atgggtatcaagttgccccgtataaaaggcaagtttaccggttgcacggt>NM_001201794_up_2000_chr2L_8382455_f chr2L:8382455-8384454ttatttatgtaggcgcccgttcccgcagccaaagcactcagaattccgggcgtgtagcgcaacgaccatctacaaggcaatattttgatcgcttgttagg...http://bioconductor.org/packages/Biostrings
Reads: FASTQ files
- The
ShortReadpackage can be used for lower-level access to FASTQ files.readFastq(),FastqStreamer(),FastqSampler()
Input & manipulation, FASTQ file example:
@ERR127302.1703 HWI-EAS350_0441:1:1:1460:19184#0/1CCTGAGTGAAGCTGATCTTGATCTACGAAGAGAGATAGATCTTGATCGTCGAGGAGATGCTGACCTTGACCT+HHGHHGHHHHHHHHDGG<GDGGE@GDGGD<?B8??ADAD<BE@EE8EGDGA3CB85*,77@>>CE?=896=:@ERR127302.1704 HWI-EAS350_0441:1:1:1460:16861#0/1GCGGTATGCTGGAAGGTGCTCGAATGGAGAGCGCCAGCGCCCCGGCGCTGAGCCGCAGCCTCAGGTCCGCCC+DE?DD>ED4>EEE>DE8EEEDE8B?EB<@3;BA79?,881B?@73;1?---#####################http://bioconductor.org/packages/ShortRead
Quality scores: 'phred-like', encoded. See http://en.wikipedia.org/wiki/FASTQ_format#Encoding
Biostrings, DNA or amino acid sequences
Classes
XString,XStringSet, e.g.,DNAString(genomes),DNAStringSet(reads)
Methods
- Manipulation, e.g.,
reverseComplement() - Summary, e.g.,
letterFrequency() - Matching, e.g.,
matchPDict(),matchPWM()
Related packages: BSgenome for working with whole genome sequences, e.g., ?"getSeq,BSgenome-method"
Aligned reads: SAM/BAM files
Input & manipulation: Rsamtools - scanBam(), BamFile()
SAM Header example
@HD VN:1.0 SO:coordinate@SQ SN:chr1 LN:249250621@SQ SN:chr10 LN:135534747@SQ SN:chr11 LN:135006516...@SQ SN:chrY LN:59373566GenomicAlignments, Aligned reads
The GenomicAlignments package is used to input reads aligned to a reference genome. See for instance the ?readGAlignments help page and vignette(package="GenomicAlignments", "summarizeOverlaps")
Classes - GenomicRanges-like behaivor
GAlignments,GAlignmentPairs,GAlignmentsList
Methods
readGAlignments(),readGAlignmentsList()- Easy to restrict input, iterate in chunks
summarizeOverlaps()
Genomic variants: VCF files
VariantAnnotation- Input and annotation of genomic variants
Classes - GenomicRanges-like behavior
VCF-- 'wide'VRanges-- 'tall'
Methods
- I/O and filtering:
readVcf(),readGeno(),readInfo(),readGT(),writeVcf(),filterVcf() - Annotation:
locateVariants()(variants overlapping ranges),predictCoding(),summarizeVariants() - SNPs:
genotypeToSnpMatrix(),snpSummary()
http://bioconductor.org/packages/VariantAnnotation
VCF-Related packages
ensemblVEP- query the Ensembl Variant Effect PredictorVariantTools- Explore, diagnose, and compare variant calls.VariantFiltering- Filtering of coding and non-coding genetic variants.h5vc- has variant calling functionality.snpStats- Classes and statistical methods for large SNP association studies.
http://bioconductor.org/packages/ensemblVEP
http://bioconductor.org/packages/VariantTools
http://bioconductor.org/packages/VariantFiltering
http://bioconductor.org/packages/h5vc
https://bioconductor.org/packages/release/bioc/html/snpStats.html
Obenchain, V, Lawrence, M, Carey, V, Gogarten, S, Shannon, P, and Morgan, M. VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics, March 28, 2014
Introduction to VariantAnnotation, http://bioconductor.org/packages/release/bioc/vignettes/ShortRead/inst/doc/Overview.pdf
Genome annotations: BED, WIG, GTF, etc. files
- The
rtracklayer'simportandexportfunctions can read in many common file types, e.g.,BED,WIG,GTF, ..., in addition to querying and navigating the UCSC genome browser. Check out the?importpage for basic usage.
Input: rtracklayer::import()
BED: range-based annotation (see http://genome.ucsc.edu/FAQ/FAQformat.html for definition of this and related formats)WIG/bigWig: dense, continuous-valued dataGTF: gene model
http://bioconductor.org/packages/rtracklayer