Working with text in Linux
Mikhail Dozmorov
Virginia Commonwealth University
02-03-2021
Text format
The most cross-platform format to share data
Typically, data is stored as field-delimited columns (think Excel). Delimiter may be tab character (".tsv" or ".txt" file extension), of comma (comma-separated values, ".csv")
Disadvantage - can be large
Solution - compression (gzipping), with tools to manipulate compressed files without uncompressing
Windows file compatability
Saving files in Windows and then trying to process them on Unix may cause issues
A common type of error comes from control characters, commonly seen as end of line characters in Windows.
To run script successfully, we need to remove these characters either by hand using
vimoremacsto edit the file, or by runningdos2unix myfile.shunix2doscommand also exists
String manipulation
RegEx - is a language for describing patterns in strings
grep - finds lines containing a pattern, and outputs them
sed - (stream editor) applies transformation rules to each line of text based on a pattern
awk - powerful text processing language
Regular expressions - everywhere

Regular expressions
| Expression | Description |
|---|---|
| [] | Matches a set. [abc] matches a, b, or c. [a-zA-Z] matches any letter. [0-9] matches any number. “^” negates a set, [^abc] matches d, e, f, etc. |
| ^ | Starting position anchor. ^abc finds lines starting with abc |
| \$ | Ending position anchor. xyz\$ finds lines ending with xyz |
| \ | Escape symbol, to find special characters. \* will find *. \n matches new line character, \t – tab character |
| * | Match the preceding element zero or more times. a*b matches ab, aab, aaab, etc. |
Extended regular expressions
| Expression | Description |
|---|---|
| ? | Matches the preceding element zero or one time. a*b matches b, ab, but not aab |
| + | Matches the preceding element one or more times. a+b matches ab, aab, etc. |
| | | OR operator. “abc|def” matches abc or def |
The grep command
- Find lines in an input file or stream that match a specific pattern you are looking for
grep "chrX" regions.bed | headchrX 41190000 41195000chrX 154020000 154025000chrX 81355000 81360000chrX 80805000 80810000chrX 88340000 88345000chrX 58420000 58425000chrX 98615000 98620000chrX 62330000 62335000chrX 153335000 153340000chrX 30660000 30665000- Result: Only lines that contain the text "chrX" (case-sensitive) anywhere in the line will be returned.
grep usage
Basic syntax: grep "pattern" <filename>, e.g., cat README.md | grep "use"
ls | grep "^[w|b]" - lists files/directorys starting with ”w” or ”b”
Use --color argument to highlight matched patterns
Fine-tuning your grep
-v - inverts the match (lines that do not contain pattern)
-i - matches case insensitively
-H - prints the matched filename
-n - prints the line number
-f
-w - forces the pattern to match an entire word (e.g., "chr1" but not "chr11")
-x - forces patterns to match the whole line
Escape special characters, e.g., grep \"gene\"
sed - stream editor
Most common usage – substitute a pattern with replacement. Basic syntax:
sed 's/pattern/replacement/'
echo "The Internet is made of dogs" | sed 's/dogs/cats/' - replaces "dogs" with "cats", so the final output is "The Internet is made of cats"
echo "dogs, dogs, dogs" | sed 's/dogs/cats/g' - global substitution with "g" modifier. The final output is "cats, cats, cats"
sed - stream editor
Special characters – escape with "\"
echo "1*2*3" | sed 's/\*/-/g’ - outputs "1-2-3"
Regular expressions – use as in grep, with "-E" argument for extended regex
echo "tic-tac-toe" | sed 's/[ia]/o/g' | sed 's/e$/c/' - outputs "toc-toc-toc"
Delete line(s) – sed 'X[,Y]d' deletes line X through Y
cat <filename> | sed '1d' - deletes first line (e.g., header)
cat <filename> | sed '10,37d' - deletes lines from 10 through 37
awk
A more traditional programming language for text processing than sed. Awk stands for the names of its authors “Alfred Aho, Peter Weinberger, and Brian Kernighan”
- Each column is referred to by number, e.g.
$1for the first column $0is referred to the whole line- Note "column" is defined as a non-contigious text. So, space- and tab-separated words are equivalent for awk
- Use
-F "\t"to override field separator, useOFS="\t"to override spaces to tabs as an output field separator awkprocess each row, and operates on column values- Commands are wrapped in single quotes
man awkfor more
Conditional output with awk
Only report annotations in
cpg.bedthat are for chromosome 1awk '$1 == "chr1"' cpg.bed# Equivalentlycat cpg.bed | awk '$1 == "chr1"'Only report annotations in
cpg.bedwhere the end coordinate is less than the start coordinate.awk '$3 < $2' cpg.bed
Special variables
- The NR (number of records) variable
Example: Report the 100th line in the file
awk 'NR == 100' cpg.bedThe NF (number of fields) variable
- Example: Report the number of tab-separated columns in the first 10 lines of
cpg.bedawk -F "\t" '{print NF}' cpg.bed | head
Impose multiple filtering criteria with the AND ("&&") operator
Report the 100th through the 200th lines in the file
awk 'NR>=100 && NR <= 200' cpg.bedReport lines if they are the 100th through the 200th lines in the file OR (||) they are from chr22
awk '(NR>=100 && NR <= 200) || $1 == "chr22"' cpg.bed
Computations in awk
Print the BED record followed by the length (end - start) of the record
$0refers to the entire input lineIf using a
printstatement, you must add curly brackets between the single quotes describing the program.Example: Prints first 3 columns, the 2nd numerical column is increased by 100, the 3rd is decreased by 100
awk '{print $1, $2+100, $3-100}' cpg.bedBy default, output is separated by a space. Prefer tabs
- BEGIN: before anything else happens, execute what is in the
BEGINstatement. Then start processing the input. - Print the BED record followed by the length (end - start) of the record. Separated by a TAB, the OFS (output field separator)awk 'BEGIN{OFS="\t"}{print $0, $3-$2}' cpg.bed# orawk '{len=($3-$2); print $0"\t"len}' cpg.bed
bioawk - awk modified for biological data
Bioawk is an extension to Brian Kernighan's awk, adding the support of several common biological data formats, including optionally gzip'ed BED, GFF, SAM, VCF, FASTA/Q and TAB-delimited formats with column names
It also adds a few built-in functions and an command line option to use TAB as the input/output delimiter.
When the new functionality is not used, bioawk is intended to behave exactly the same as the original BWK awk.
https://github.com/vsbuffalo/bioawk-tutorial
https://github.com/ialbert/bioawk/blob/master/README.bio.rst
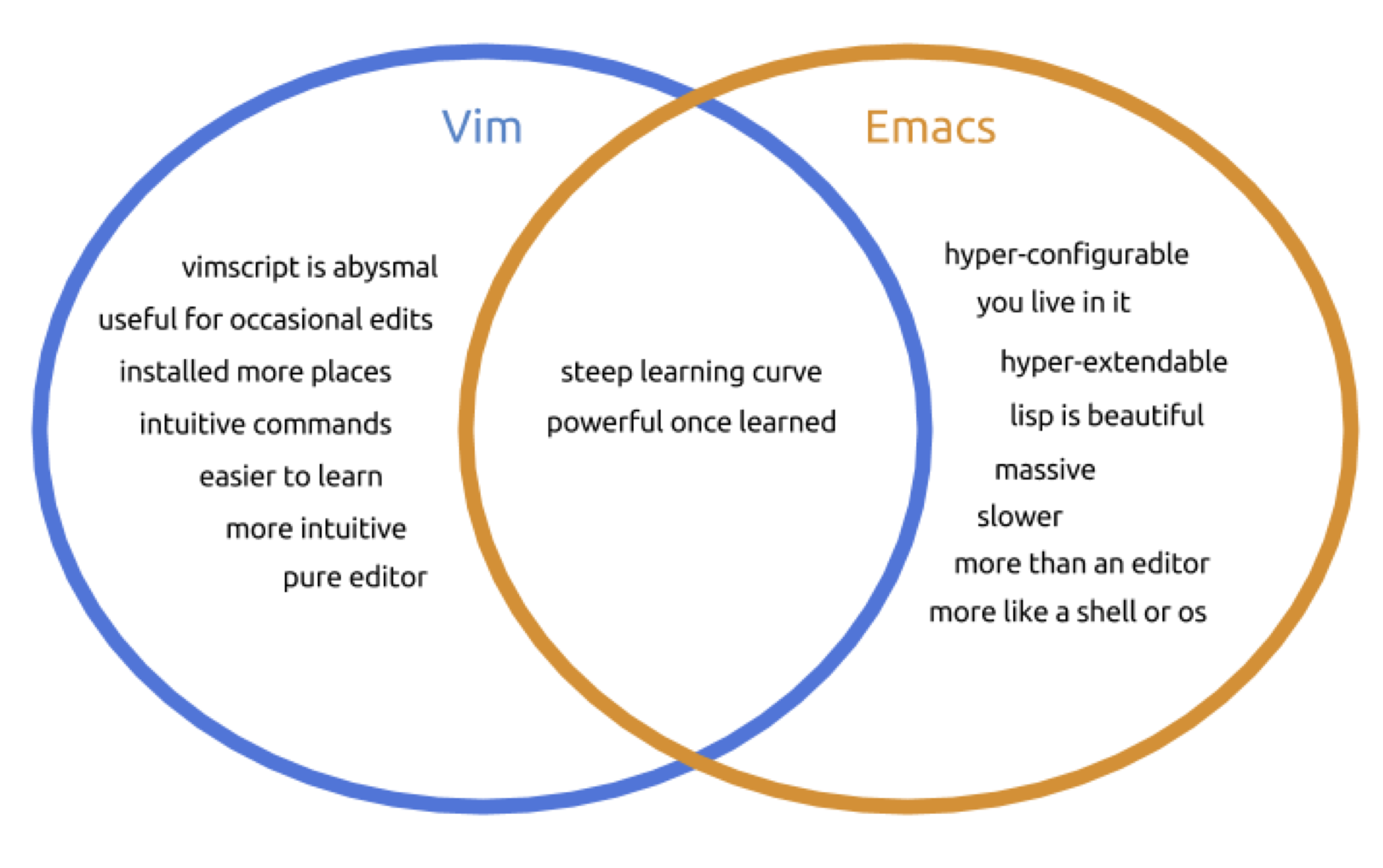
Command-line text editor
nano- simple editorvim- Created by Bill Joy, 1976. Advantages: Supremely intuitive once basics are learnedemacs- Created by Richard Stallman, 1976. Advantages: Unparalleled power and configuration
vim basics
Start vim on a file: vim <filename>
Keyboard shortcuts for two modes:
i- editor mode, to typeEsc- command mode. Press “:” and enter a command
Important keyboard shortcuts:
:w- write changes:wq- write changes and quit:q!- force quit and ignore changes
Basic vim commands
k, j, l, h, or arrows - navigation
v - (visually) select characters
V (shift-v) - (visually) select whole lines
d - cut (delete) into clipboard
dd - cut the whole line
y - copy (yank) into clipboard
P (shift-p) - paste from clipboard
u - undo
Find and replace in vim
In command mode:
/pattern- search for pattern, “n” – next instance:s/pattern/replacement/g- search and replace
:help tutor - learn more vim
References
- Regular expression, Unix commands, Python quick reference, SQL reference card. http://practicalcomputing.org/files/PCfB_Appendices.pdf